Research
Recently, the field of immune regulation has become more expansive and tremendously complex. The balance between immunity and tolerance determines the outcome of many human diseases such as infection, autoimmunity and cancer. During the last decade a unique active mechanism of immune suppression by a dedicated population of so-called regulatory T (Treg) cells has become the focus of intensive investigation. The understandings of molecular suppressor function of Treg cells and the cellular interaction between Treg cells and other immune cell populations lie at the heart of immune regulation. Recent advances have made Treg biology a fascinating research field amenable to mechanistic dissection. Below, I outline the ongoing and future research projects in the lab that aim to identify the molecular determinants of differentiation and function of Treg cells. Ultimately, we hope to gain insight into the complex biology of these cells, including the maintenance of immunological tolerance to “self” and the regulation of immune responses to pathogens, commensals, and tumors.
1. Identification of miRNAs crucial for mediating Treg cell biology in healthy and diseased mice
Several thousands genes are differentially expressed in Foxp3+ Treg cells, including a subset of non-coding small regulatory microRNA (miRNA), some of which are directly regulated by Foxp3 suggesting a role for miRNA-mediated regulation of gene expression in Treg cell differentiation, maintenance or function. Catastrophic consequences and the development of complex disease phenotypes in mice harboring Treg cells devoid of miRNA suggested miRNAs control multiple facets of Treg cell biology (Fig.1).

Fig.1) miRNA deficient Treg cells are impaired in homeostasis and function.
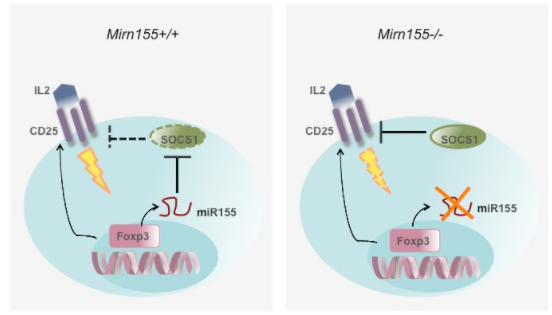
Fig.2) miR155 confers Treg cell competitive fitness.
and miR-146a (Fig.3). Currently, in addition to two aforementioned miRNAs, we have established several mouse models with deletion or overexpression of miRNAs over-represented or under-represented in Treg cells. By employing genetic, biochemical, immunological approaches and whole animal experimentation, we aim to identify the function role of individual miRNA in Treg cells vs effector T cell populations. Moreover, we are interested in further characterizing miRNA expression profiles in Treg cells isolated from different tissues in unmanipulated mice as well as in mice suffering different pathologies. We want to understand how miRNAs themselves are regulated in response to different environmental cues. Ultimately, we hope to elucidate how such regulation of miRNA expression impacts their respective targets and how the dynamic miRNA-dependent regulation enforces Treg cell functionality in diverse disease settings.
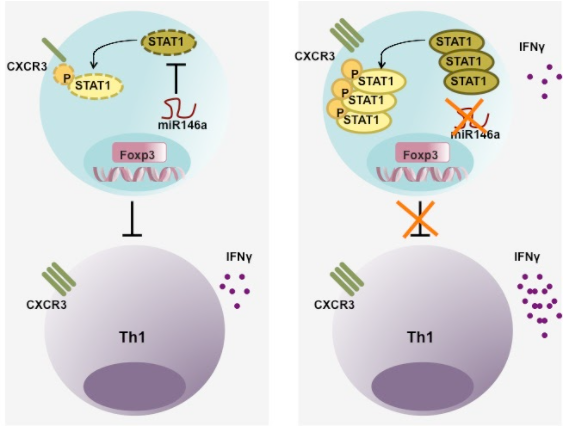
Fig.3) miR-146a maintains Treg cell identity and their suppression function in responses to type I inflammation.
2. Elucidating molecular and cellular mechanisms by which Treg cells employ to control type I inflammation
Recent studies revealed that the expression of Th lineage-specific transcription factors in Treg cells is crucial for Treg cell-mediated control of corresponding immune responses in particular tissue and inflammatory settings. Our analysis of the role of miR-146a and SOCS1 in Treg cell-dependent Th1 regulation have further extended this concept and demonstrated that both lack of Stat1 and unrestrained Stat1 activation in Treg cells would result in a similar breakdown of immunologic tolerance (Fig.4). These results suggest that Treg cell-mediated suppression of a given Th-specific immunopathology is depending upon an optimal range of activation in corresponding Th-lineage specific transcription factors. One fundamental question that remained is how Treg cells control Th1 responses. At the molecular level, we are interested in identifying important effector molecules and signalling pathways underlying Treg cell-mediated Th1 regulation. At the cellular level, we hope to understand the complex interplay between Treg cells, effector T cells and other immune cells in the surrounding environment specifically in a type I inflammatory setting. Moreover, IFNg-mediated Th1 responses and Treg cells have been associated with inflammation in many clinical settings such as infection (e.g., T. gondii), autoimmune diseases (e.g., multiple sclerosis and rheumatoid arthritis), tumors, organ transplantation and tumor. Through our study, we hope to establish the foundation for the future development of means to manipulating Treg and T effector cell function for better treatment in human patients.

Fig.4) The level of Stat1 activation in Treg cells dictates their ability to control Th1 responses.


